Detecting Kidney Diseases with Deep Learning: A CNN Approach
Medical imaging is powerful, but manually analyzing kidney scans is slow, error-prone, and exhausting for radiologists. What if a machine could automatically classify kidney conditions with high accuracy? That’s exactly what this project tackles using Convolutional Neural Networks (CNNs).
The Problem
Kidney diseases can be subtle and complex. Misdiagnosis or delayed detection can have serious consequences. Our goal: build a CNN model that automatically identifies four types of kidney conditions from images:
- Cyst – fluid-filled sacs
- Stone – mineral deposits
- Tumor – abnormal tissue growth
- Normal Kidney – healthy kidneys
By automating classification, we can assist medical professionals and speed up diagnostic workflows.
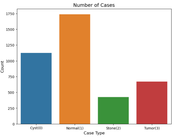
Exploring the Dataset
The dataset contains labeled images for all four categories. Here’s a quick look at the classes:
Each image is unique, but you can clearly spot differences between cysts, stones, tumors, and normal kidneys. That visual distinction is key for the CNN to learn meaningful features.
Preprocessing the Images
Raw medical images aren’t ready for deep learning. We apply a structured preprocessing pipeline:
- Resizing – standardizing image dimensions
- Normalization – scaling pixel values between 0–1
- Label Encoding – converting categories into numerical format
- Train–Validation Split – ensuring the model generalizes well
Here’s how the pipeline transforms the images:
Consistency in preprocessing is non-negotiable—any variation directly impacts accuracy.
Building the CNN Model
The CNN is the heart of this project. It’s designed to extract hierarchical patterns from kidney scans.
Key components:
- Convolutional layers with ReLU activation for feature extraction
- MaxPooling layers for spatial reduction
- Flattening layer to convert 2D features into 1D
- Dense layers for classification
- Softmax output layer for 4-class prediction
Visual summary:
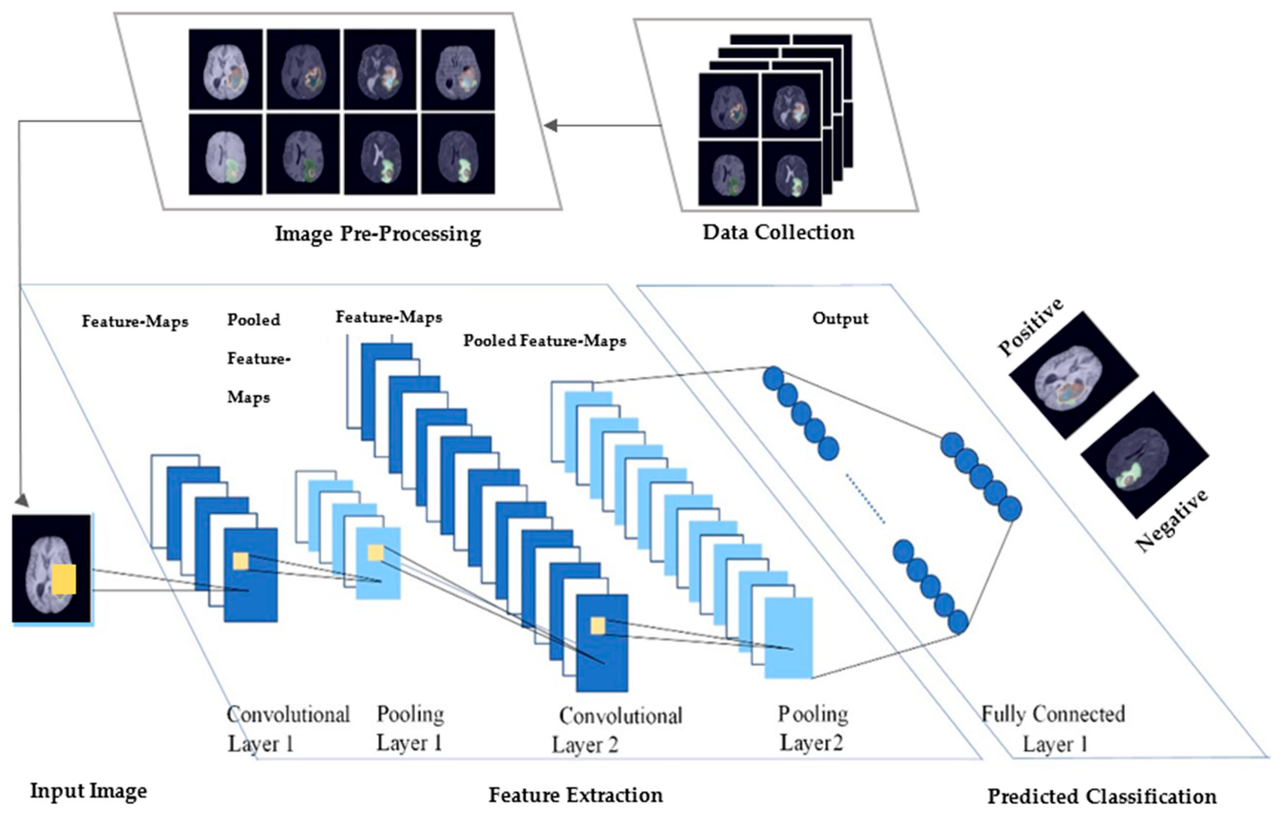
This architecture is lightweight yet effective—perfect for a first-pass medical image classifier.
Training the Model
We compile the CNN with:
- Loss:
categorical_crossentropy - Optimizer:
Adam - Metric:
accuracy
Training involves iterating over the dataset in batches, tracking both training and validation performance:

These curves show how the model converges, and help identify overfitting early.
Evaluating Performance
After training, the model is tested on unseen images. The results are impressive:
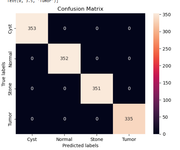
It classifies cysts, stones, tumors, and normal kidneys with strong accuracy, confirming the CNN has learned meaningful patterns.
Making Predictions on New Images
Deploying the model is straightforward:
- Resize and normalize the new image
- Feed it into the trained CNN
- Generate softmax probabilities
- Pick the class with the highest probability
A simple pipeline, but it works reliably for real-world usage.
Why Jupyter Notebook?
Jupyter Notebooks make this project easier to explore:
- Inline visualizations of kidney images
- Immediate feedback on code and model performance
- Seamless experimentation with preprocessing and architecture
For deep learning prototyping, this environment is essential.
Conclusion
This project demonstrates that CNNs can effectively classify multiple kidney conditions from images. By handling cysts, stones, tumors, and healthy kidneys, we can support faster, automated, and accurate diagnostics.
Automated medical imaging isn’t just futuristic—it’s practical, impactful, and ready to assist healthcare professionals today.
